 Tandem repeats (TRs) are short sequences of DNA repeated over and over again like the DNA letter sequence TACTACTAC, which is a repetition of TAC three times (Figure 1). In the early days of genomics, these tandem repeats were originally designated as nonfunctional or junk DNA. With the exception of a few cases of tandem repeats being involved in human disease, this type of repeat variation was often believed to be neutral in its effect on the organism.
Tandem repeats (TRs) are short sequences of DNA repeated over and over again like the DNA letter sequence TACTACTAC, which is a repetition of TAC three times (Figure 1). In the early days of genomics, these tandem repeats were originally designated as nonfunctional or junk DNA. With the exception of a few cases of tandem repeats being involved in human disease, this type of repeat variation was often believed to be neutral in its effect on the organism.
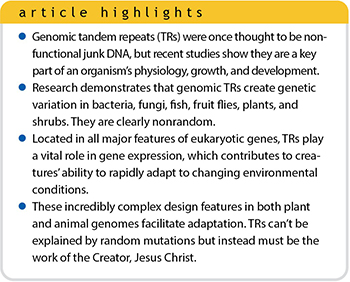
However, over the past several decades scientists have shown that TRs are functional and designed DNA features. And contrary to initial declarations of them being nonfunctional, researchers have found that TRs and their variability in length have useful consequences in the biology, growth, and development of an organism toward adaptive outcomes.
There are two different types of TRs.1 One type called a microsatellite consists of repeats in which the basic repeated unit consists of one to nine DNA letters (nucleotides). The second type is called minisatellites and consists of units that are greater than nine nucleotides (see Figure 1).

The variability in TRs consists in different lengths of the repeats such that one variation might contain 12 repeat units while another might have 13. Repeat variability can exist within the same genome on homologous chromosomes (one from the father and one from the mother) or between individuals of the same created kind, allowing for adaptive variation within a population.
The mechanisms for altering repeat lengths of a TR are still largely unknown, although there has been speculation concerning how this might occur. Some researchers have suggested it occurs during DNA replication and cell division or recombination, which takes place during meiosis (production of sperm and egg cells).1,2
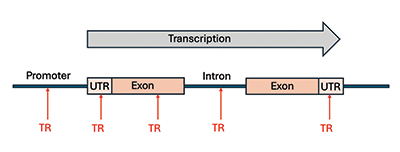
The mere fact that TRs are a ubiquitous feature of genes points to their functionality and importance. The region at the beginning of a gene is called a promoter (gene regulatory feature). It has been determined that as many as 10% to 20% of eukaryotic gene promoters (found in creatures with a cell nucleus) contain TRs that function as regulatory switches.3 But before we can delve into the phenomena of TRs in genes, we need to review how a gene is structured in a eukaryotic genome.4,5
First, it is important to understand that genes are in pieces. They contain regions called exons that code for a protein or functional RNAs (Figure 2) with intervening sections called introns between exons that do not code for proteins or functional RNAs. The introns contain many important signals and features but are spliced out of the RNA copy of the gene (messenger RNA or transcript). As noted above, regulatory TRs can be found in gene promoters, but they can also be found throughout the other parts of the gene (Figure 2).
Nonrandom Adaptive TRs in Gene-Coding Sequences
The first evidence that TRs cause adaptive and useful variation in the protein-coding regions of genes was found in several different types of bacteria in which the variable-length TRs would switch off or on the production of a cell membrane protein.3 This in turn had significant effects on other proteins in the cell membrane. The end result was that the bacterial populations contained what was termed phase variation that facilitated rapid adaptation to a broad range of environmental conditions.
In another, more complex eukaryotic single-cell microbe, brewer’s yeast, TRs are found in up to 22% of the coding regions of all genes.3 And like bacteria, these variable-length TRs create adaptive population diversity that allows them to rapidly adjust to the conditions around them, e.g., generating dynamic protein interactions on the yeast’s cell surface for optimized air-liquid interfaces. Other common yeast TRs occur in regulatory proteins (transcription factors) that have a broad effect upon the cell’s genetic networks and circuits and even affect the epigenetic states of chromosomes.
Circadian clock systems function in creatures to keep cells, organs, and the whole organism in sync during the day/night 24-hour cycle.6,7 In multicellular eukaryotes, variable-length TRs in coding regions have been shown to act as fine-tuning mechanisms for circadian clocks.3
Proteins are chains of amino acids. In the fungus Neurospora crassa, a gene encoding a transcription factor named White Collar-1 (WC-1) has a TR tract that codes for a repetitive string of glutamine amino acids in the WC-1 protein. Its variability in length acts like a fine-tuning knob. The population variability in the TR length enables the fungus to rapidly adjust its circadian clock to the environmental inputs of both the light cycle and temperature.
In the fruit fly, the per (period) gene has a six-base repeat that codes for a repetitive sequence of two amino acids, threonine and glycine. The two most common TR length variants in fly populations have either 17 or 20 repeats. The 17-repeat TR enables the flies to adjust their circadian clock under cold temperatures, while the 20-repeat TR allows clock adjustment under warm temperatures.
And a recent study showed that polyglutamine repeats in regulatory proteins derived from a TR of cytosine-adenine-guanine in two different genes (CLOCK and BMAL1) help regulate circadian clocks in several species of pelagic seabirds. This confers fine-tuned local adaptation to latitude and helps adjust breeding time.8
Nonrandom Adaptive TRs in Regulatory Sequences
In addition to gene promoters that regulate gene expression, another important type of gene regulatory sequence is called an enhancer element, which can be hundreds of bases away from the gene or even embedded inside the gene. Both promoters and enhancers are activated by DNA-binding proteins called transcription factors that bind to specified target sequences within them. In these types of regulatory sequences, TRs have been found to play crucial roles in a variety of biological processes with functional implications.9 Certain TRs in promoters and enhancers themselves regulate gene expression by acting as binding sites for transcription factors.

When TRs occur in gene promoters, the variability in repeat length has been associated with a wide host of traits related to adaptability. In tilapia, an important aquacultural fish, variable repeats of CA have been found to occur in the promoter of an osmoregulatory gene called PRL1.3 These TR variations confer a range of adaptability to water salinity. These variations are also associated with the size of the fish.
Plants and Their TR-Based Adaption
Plants are tethered to the soil (sessile) and cannot get up and move to places that might be more favorable with fewer environmental challenges. As a result, they rely heavily on robust adaptive systems to prosper where they are planted, and their copious adaptive use of TRs in the genome is no exception.

In a recent study of the small weedy plant Arabidopsis, researchers analyzed many TRs in the genome, revealing that the TRs clustered near genes were involved in development, stress responses, and plant hormone pathways.10 The data also demonstrated that the closer a TR is to a gene, the more likely it is to influence expression of that gene.
A surprising discovery was that TR sites in protein-coding sequences that encode repeated amino acids, as discussed above, also affect gene expression. For one protein in particular, it was experimentally verified that TR variation in the protein-coding region also created variation in the binding of transcription factors in the gene’s promoter. Additionally, TR variation was found to be associated with multiple adaptive traits including root morphology, biotic stresses, abiotic stresses, and plant cell immune receptors.

Another recent study was done with the Siberian peashrub Caragana, which is widely known for its remarkable adaptability to harsh environments across Siberia and China.11 Caragana plants have very distinctive geographical distribution patterns that reflect a wide variety of adaptive trait variations in plant morphology, physiology, and biochemistry. This amazing array of innate biological diversity has made Caragana an ideal plant system for studying how TR variability may play a role in adaptation.
In this particular study, researchers identified variable TRs in a large number of genes in both coding and noncoding regions. They employed a transcriptome—the set of all RNA transcripts present in a cell—to analyze gene expression across the whole genome for 12 different Caragana species across diverse locations in China. Their result was a large database of TR variations connected to active genes that were significantly linked to functional traits related to climate, altitude, and soil conditions (salinity and water availability).
The specific conclusion was that the variation of 264 gene-based TRs significantly controlled the expression of the genes in which they were located. Additionally, another 2,424 TRs were located in genes that had variable expression amongst Caragana species. Statistically, the expression of these genes was correlated with 19 different environmental adaptations and 16 different functional traits across the various habitats.
Conclusion
Until the past several decades, TRs were considered to be nonfunctional DNA that had no purpose—so-called “junk DNA.”12 While a number of variable-length TRs were initially shown to be connected to some heritable human diseases, a broad range of new studies discussed in this article are showing that TRs are clearly important design features of the genome that are involved in animal and plant adaptation.
Instead of being leftover evolutionary junk DNA, TRs demonstrate precise function and intricate engineering that could never arise through random means or natural processes. Once again, such a clever genetic mechanism points to the all-wise Creator, the Lord Jesus Christ.
References
- Gemayel, Ρ. et al. 2012. Beyond Junk-Variable Tandem Repeats as Facilitators of Rapid Evolution of Regulatory and Coding Sequences. Genes. 3 (3): 461–480.
- For more information on meiosis and recombination, see Tomkins, J. P. 2024. Genetic Recombination: A Regulated and Designed Chromosomal System. Acts & Facts. 53 (4): 16–19.
- Gemayel, Ρ. et al. 2010. Variable Tandem Repeats Accelerate Evolution of Coding and Regulatory Sequences. Annual Reviews of Genetics. 44: 445–477.
- Tomkins, J. P. 2012. The Irreducibly Complex Genome: Designed from the Beginning. Acts & Facts. 41 (3): 6.
- Tomkins, J. P. 2014. Gene Complexity Eludes a Simple Definition. Acts & Facts. 43 (6): 9.
- Tomkins, J. P. 2016. Circadian Clocks, Genes, and Rhythm. Acts & Facts. 45 (7): 14.
- Tomkins, J. P. A Time for Everything – Your Body’s Internal Clock. Answers Magazine. Posted on answersingenesis.org August 25, 2018.
- Nirchard, K. et al. 2023. Circadian Gene Variation in Relation to Breeding Season and Latitude in Allochronic Populations of Two Pelagic Seabird Species Complexes. Scientific Reports. 13 (1).
- Liao, X. et al. 2023. Repetitive DNA Sequence Detection and Its Role in the Human Genome. Communications Biology. 6 (1), article 954.
- Reinar, W. B. et al. 2021. Length Variation in Short Tandem Repeats Affects Gene Expression in Natural Populations of Arabidopsis thaliana. Plant Cell. 33 (7): 2221–2234.
- Wanf, Q. et. al. 2024. The Potential Role of Genic-SSRs in Driving Ecological Adaptation Diversity in Caragana Plants. International Journal of Molecular Sciences. 25 (4).
- Tomkins, J. P. Human Genome 20th Anniversary…Junk DNA Hits the Trash. Creation Science Update. Posted on ICR.org April 12, 2021.
Dr. Tomkins is a research scientist at the Institute for Creation Research and earned his Ph.D. in genetics from Clemson University.