The ICR life sciences team has identified several key research questions in the field of origins biology. Our current task is reviewing the literature concerning the evolutionary tree of life,1 with a focus on papers related to molecular data.2,3 Specifically, we are reexamining the raw data and looking for evidence of discontinuity.3
To compare species at the molecular level, the DNA or protein sequences in these species must be obtained. These data are derived from two sources: 1) New lab experiments done by the investigator, and 2) publicly available data from already published experiments. The latter represents a rich resource for comparisons since much published biological sequence data already exist. Together, these two sources comprise the core of the data used for species comparisons.
Once the DNA or protein sequences of various species are obtained, there are many ways to compare these species molecularly. One of the more common ways is based on the gene sequence differences among creatures. Since diverse organisms often share a core set of genes, and since the sequences of these shared genes often differ significantly (depending on the gene), creatures can be distinguished based on their unique gene sequences. Quantifying the differences in sequence among species is a powerful way to compare them.
A variety of methods exists by which gene sequence differences can be measured across species. One method we have been employing is the CLUSTAL W software program, which is freely available online through the European Bioinformatics Institute (EBI). This program aligns a given set of input sequences across the entire length of each sequence and seeks to optimize the alignment based on the input parameters given to the program. When comparing multiple sequences at once, the EBI version of CLUSTAL W first performs pair-wise comparisons (the alignment of all sequences pair by pair, one pair at a time) before building an alignment of all input sequences. After the alignment has been optimized, one of the parameters CLUSTAL W reports is the percent identity between pairs of input sequences. This parameter is a direct report of the sequence differences between two creatures. By performing pair-wise comparisons across multiple species, we can categorize species based on their sequence differences from all other species.
A hypothetical example of the results of a CLUSTAL W alignment is shown below. The numbers in individual boxes depict the individual pair-wise comparisons of the species listed at the beginning of the column and row; these numbers represent percent identity between the gene sequences from the two species compared. In this hypothetical 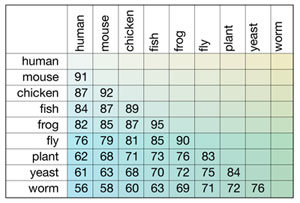 table, each creature shows a hierarchy of percent identity among other creatures. This type of output is useful for classifying and establishing relationships among creatures.
table, each creature shows a hierarchy of percent identity among other creatures. This type of output is useful for classifying and establishing relationships among creatures.
In sum, molecular sequence alignments are a very useful way to analyze and categorize differences among diverse creatures, and we anticipate that such comparisons will manifest the gaps in nature that Genesis 1 predicts.
Previous molecular comparisons among creatures have revealed differences between species that Darwin never anticipated. Read about these fascinating results in upcoming issues of Acts & Facts!
References
- Jeanson, N. 2010. Literature Review: Simplifying the Research Process. Acts & Facts. 39 (11): 6.
- Jeanson, N. 2010. New Frontiers in Animal Classification. Acts & Facts. 39 (5): 6.
- Jeanson, N. 2010. Literature Review: Molecular Data and the Tree of Life. Acts & Facts. 39 (12): 6.
* Dr. Jeanson is Research Associate and received his Ph.D. in Cell and Developmental Biology from Harvard University.
Cite this article: Jeanson, N. 2011. Looking for Molecular Discontinuity: The Use of Sequence Alignments. Acts & Facts. 40 (1): 6.