 The field of epigenetics is one of the most exciting and rapidly expanding scientific research areas in the study of the genome and how it responds adaptively in organisms. The term epigenetics is derived from genetics plus the Greek prefix epi, which means “on top of” or “in addition to.” In other words, it’s an additional type of genomic language that overlays the DNA code that controls how genes are switched on or off. It even determines the three-dimensional structure of the chromosomes in the cell’s nucleus.
The field of epigenetics is one of the most exciting and rapidly expanding scientific research areas in the study of the genome and how it responds adaptively in organisms. The term epigenetics is derived from genetics plus the Greek prefix epi, which means “on top of” or “in addition to.” In other words, it’s an additional type of genomic language that overlays the DNA code that controls how genes are switched on or off. It even determines the three-dimensional structure of the chromosomes in the cell’s nucleus.
Epigenetic changes in the genome are modulated dynamically according to sensory input that the body detects from its physical surroundings, signaling molecules it receives from other creatures (e.g., gut microbes), diet, and even stress. In fact, many epigenetic changes can be heritable and affect traits passed along to children and grandchildren. In many animals and plants, these heritable changes also prime the offspring to be specifically adapted to some aspect of the environment—giving them a preconditioned jumpstart on life.
Two different systems can epigenetically modify an organism’s chromosomes. The first and most easily studied system is known as cytosine methylation, which is the addition of chemical methyl groups to the actual DNA base molecules or nucleotide letters. The second system is the modification of proteins called histones that integrate with the DNA and allow it to be packaged and spatially structured in different ways. Both of these types of epigenetic modification determine the accessibility of DNA to protein regulators called transcription factors that bind to regulatory sites in the DNA and control gene activity. This article will discuss each of these modification systems and then show how our current knowledge of epigenetics is connected to creaturely adaptation.
Cytosine Methylation
The DNA code comprises the ordered sequence of the nucleotide letters A (adenine), C (cytosine), T (tyrosine), and G (guanine).1,2 Epigenetic modification of the actual DNA molecule occurs specifically on the cytosines by the addition of small epigenetic tags called methyl groups—hence the term cytosine methylation. This type of modification is especially pronounced in regions of the genome that have not only a high C content but Gs as well, since Cs pair with Gs in the double-stranded DNA molecule. The presence of these methyl tags in the DNA molecule plays a major role in the way that genes are expressed, i.e., turned off and on.
The DNA sequence of an organism is generally the same across the creature’s body in the various different tissues. However, the methylation status or profile across the genome will vary dynamically depending on what kind of tissue the cell is located in, such as heart, lung, bone, or brain tissue. In fact, the methylation states can even vary across cells within the same tissue, since many organs are composed of different types of cells. In humans, greater than 4% of the cytosines in the three-billion-base genome are methylated. In the regions where there are dense amounts of cytosines, greater than 80% are methylated.2
In general, when the control regions of genes known as promoters are heavily methylated, the gene is turned down like an adjustable light switch being dimmed. In addition, many other switches called enhancer elements that are around the genome outside of genes can also become methylated, which regulates the level of gene activity as well. But not only are these regulatory switches regulated by DNA methylation, but the main body of genes following the promoter are also subject to different levels of methylation, which has an effect on the activity of that particular gene. The more methylated the DNA is at the start of a gene region, the less active the gene generally is.
The methylation state of DNA across the genome is highly controlled, altered, and maintained by specialized machinery called readers, writers, and erasers.2 The readers monitor the epigenetic state of DNA methylation across the genome. The writers attach methyl groups to Cs dynamically according to the needs of the cell and the tissue in which it is located. The erasers remove the methyl groups across the genome also according to the cell’s demands. The end result is that these actions change and regulate gene expression.
The addition of methyl tags in the writing process is done by enzymes called DNA methyltransferases.2 In humans and other mammals, scientists have discovered at least three different methyltransferases that function as writers. The erasing function of methyl tags is done by enzymes called demethylases, which remove methylcytosines and replace them with regular cytosines using specialized enzymatic machinery. The exact cellular machinery of specified demethylation remains much less understood than that of methylation. In addition, demethylation can also occur when the DNA is replicated during the process of cell division (mitosis), a process known as passive demethylation.
While the DNA code is very similar in all cells throughout the human body, the epigenetic code and its patterns vary depending on cell and tissue type. The specific genome-wide epigenetic profile in a particular cell type as it relates to these cytosine tags is called the methylome. In its relation to human health and disease, much research has attempted to find aberrant methylome profiles in certain cell types relating to things like cancer, diabetes, and heart disease. Furthermore, methylome profiles are radically different between various types of organisms. For example, DNA methylation in insects is mostly within gene bodies (coding region of the gene), whereas in plants and mammals it’s mostly within a class of DNA called transposable elements.3
Histone Modifications
Nearly every cell in the human body has the same genetic code contained within approximately two linear meters of DNA. This huge amount of DNA in the nucleus of a cell presents a considerable organizational and functional engineering challenge. The DNA must be packaged within a microscopically small space while also being dynamically accessible to a host of specialized genetic machinery in a spatially and temporally coordinated manner.
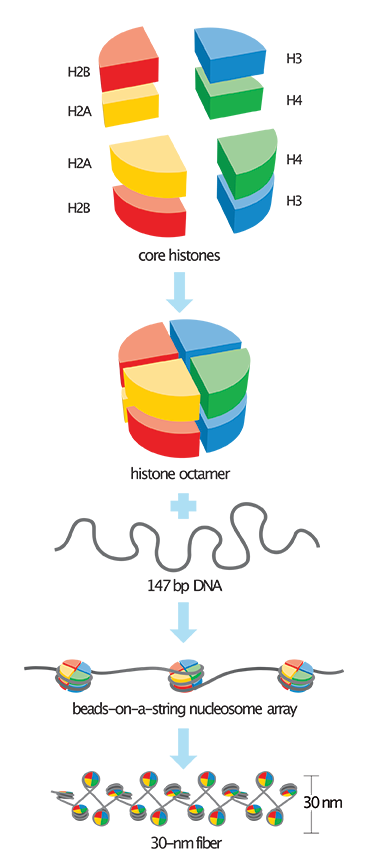
The divinely engineered solution is achieved by the DNA molecules being spooled around a specialized structure containing two pairs of four different proteins called histones.4 These proteins are designated H2A, H2B, H3, and H4, and form a spool-shaped structure that allows for 147 bases of DNA to be wrapped around it to form a bead called a nucleosome (Figure 1). In between each nucleosome is a small stretch of DNA called linker DNA that’s about 10 to 80 nucleotides long, depending on the organism. This whole packaging layout is often referred to as the beads-on-a-string model.
The DNA and histones that together form the basic structure of chromosomes is collectively called chromatin. This chromatinized DNA can also be progressively folded into more compact and condensed chromosomal structures. In general, the more folded and packaged DNA is along a chromosome, the less gene activity there is in that particular region. The more decondensed and even depleted of nucleosomes the DNA is, the more genetically active it is. In fact, DNA templated processes such as transcription (making an RNA copy), recombination (DNA region exchange and shuffling during meiosis), DNA replication, or DNA repair rely on unpackaged “open” chromatin.4
Histones have the ability to be modified in highly specific ways, which is another important feature of nucleosomes in the accessibility of DNA to various chromatin processes such as gene activity. Each histone protein has a tail that sticks out that can be modified with specialized tags.4 These histone tail modifications cause the DNA to become more or less accessible to the machinery involved in gene expression.
These protein modifications are called post-translational because they occur after the histone proteins are made. The modifications include methylation, acetylation, phosphorylation, sumoylation, and ubiquitylation. Because these different tags can be added in different ways to eight of the different histone proteins in the nucleosome, a language of over 100 different histone character states can be achieved, leading to a very complex system of chromatin regulation. In addition, the combination of histone modifications also interacts with the system of cytosine methylation in the DNA discussed above, thus controlling the activity and access of the DNA methylases.
Current research in chromatin structure points toward a continuously and dynamically changing genomic architectural landscape. In other words, chromatin is continuously morphing and interconverting between various states of accessibility and three-dimensional structure. Thus, chromatin and its nucleosomes represent much more than a merely static and inert packaging structure. In fact, the genome as a whole is a dynamic scaffold that’s capable of responding to specific cues that regulate the accessibility of DNA to various systems and components of the cellular machinery in the nucleus.
Epigenetics in Adaptation
One of the main ways epigenetics facilitates adaptation is by providing altered and heritable gene expression that increases the adaptability of a creature’s offspring to novel environments or conditions.5 In other words, different heritable adaptive traits can be produced from the same genetic background depending on prior events that lead to heritable epigenetic modifications.
Epigenetic inheritance is especially important for plants because they can’t get up and move around to find a more favorable environment. In this respect, one of the first studies highlighting heritable epigenetic adaptation was in the small weedy plant Arabidopsis (thale cress). The study was so novel and anti-Darwinian that the paper was titled “Evolution heresy? Epigenetics underlies heritable plant traits.”6
In this study, researchers tested 80 different Arabidopsis strains that were nearly genetically identical except for some that lacked a gene controlling proper DNA methylation patterns. Thus, the test focused on a large population of genetically similar plants that had both normal and aberrant levels of methylation in their genomes. The researchers tested the plants over several generations for flowering time and root growth. The goal of the study was to determine if variability in these traits was passed along from generation to generation by genetic or epigenetic differences. They found that the DNA sequence in the regions of the Arabidopsis genome that control both flowering time and root length was identical for all 80 plants and did not contribute the observed variability. What they discovered was that the inherited variability for these important traits was associated with cytosine methylation changes.
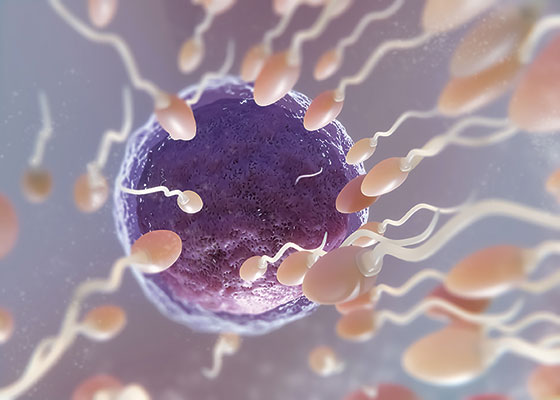
In another experiment with animals, researchers conditioned male mice to fear a cherry blossom-like scent called acetophenone by giving them electric shocks every time it was pumped into their cages.7 After 10 days of this treatment, whenever the cherry blossom scent was in the air, even without an electric shock the mice went into panic mode, expecting to get electrocuted. The researchers also found that these mice developed more smell receptors specifically associated with the scent, which allowed them to detect the acetophenone chemical at lower concentrations.
When the researchers examined the sperm of the males that were shock-conditioned, they found that the odorant receptor gene responsible for acetophenone detection was cytosine-methylated differently compared to the same gene in control mice. In fact, the gene had much less methylation, indicating that it was upregulated to create more odorant receptors. The researchers then inseminated females with the traumatized mice’s sperm. The baby mice, who had never interreacted with their fathers, had more acetophenone odorant receptors and also became agitated when acetophenone filled the air. Amazingly, this same trait was epigenetically passed on for several generations.
While these two examples in plants and mice documented adaptive epigenetic inheritance related to cytosine methylation, adaptive histone modifications are also being reported in several different types of creatures. In both fission and budding yeast, up to 20 generations can heritably maintain a variety of specifically induced histone modifications that were regulated by environmental conditions in the yeast’s culture medium.5 In the fruit fly (Drosophila), histone modifications were found to be modified related to temperature changes or starvation.5 And in the small roundworm (Caenorhabditis elegans), histone-related gene expression patterns were found to be generationally passed on in response to both heat stress and exposure to bacterial pathogens.5
Conclusion
Discoveries in epigenetics present a variety of substantial problems for evolution’s failed mutation-selection paradigm. If random genetic alterations of the DNA sequence are not connected with adaptative traits, then nature as a mythical selective agent has nothing to work with.
In fact, the reality of the scientific evidence is that creatures contain innate systems of epigenetic modification as an engineered adaptive response. First, the methylation of DNA and histone modifications are not random features in the genome. These purposeful chemical tags are placed at specific chromatin addresses all over the genome in response to various environmental cues.
Second, complex cellular machinery and surveillance systems must interpret the huge diversity of epigenetic tags not only according to the creature’s environment but also based on the type of cell and tissue in which the chromatin is located. Third, for the complete epigenetic system to be passed along during cell growth and to the next generation, there exists yet another separate and necessary system that copies the chromatin profile when the cells are replicated or sperm and egg are produced for reproduction.
The evolutionary story of random mutations and natural selection can’t account for DNA’s exquisitely engineered systems. Only our Creator Jesus Christ can.
References
- Kumar, S., V. Chinnusamy, and T. Mohapatra. 2018. Epigenetics of modified DNA bases: 5-methylcytosine and beyond. Frontiers in Genetics. 9: 640.
- Mattei, A. L., N. Bailly, and A. Meissner. 2022. DNA methylation: A historical perspective. Trends in Genetics. 38 (7): 676-707.
- Ashe, A., V. Colot, and B. P. Oldroyd. 2021. How does epigenetics influence the course of evolution? Philosophical Transactions of the Royal Society B: Biological Sciences. 376 (1826): 20200111.
- Millán-Zambrano, G. et al. 2022. Histone post-translational modifications—cause and consequence of genome function. Nature Reviews Genetics. 23: 563-580.
- Stajic, D. and L. E. T. Jansen. 2021. Empirical evidence for epigenetic inheritance driving evolutionary adaptation. Philosophical Transactions of the Royal Society B: Biological Sciences. 376 (1826): 20200121.
- Pennisi, E. 2013. Evolution heresy? Epigenetics underlies heritable plant traits. Science. 341 (6150): 1055.
- Dias, B. G. and K. J. Ressler. 2014. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nature Neuroscience. 17: 89-96.
* Dr. Tomkins is Research Scientist at the Institute for Creation Research and earned his Ph.D. in genetics from Clemson University.



