Scientists have long been baffled as to what actually tells proteins called transcription factors (TFs) where to bind in the genome to turn genes off and on. However, new research incorporating the three-dimensional shape of DNA has revealed an incredibly complex system of interacting biochemical codes.1
We know that genes are turned off and on across the genome by intricate networks of transcription factors which bind to DNA in strategic places in and around the genes. But discovering what tells the transcription factors where to bind has proven extremely difficult.
Traditionally, researchers have focused on finding out which DNA sequences bind with which transcription factors, thinking there may be a consistent pattern. However, this approach has proven only mildly successful because any given transcription factor will bind to a diversity of different DNA sequences. This is despite the fact that their DNA binding is highly tissue specific and tightly regulated. In fact, the authors of the current paper stated, "Underlying mechanisms that explain the highly specific binding of TFs to their genomic target sites are poorly understood."
But the new research has revealed that two main things influence how DNA is read by transcription factors:
- the actual sequence of the DNA and,
- the conformational shape of the DNA as it's being read.
The sequence of these binding sites has been fairly easy to determine by sequencing the fragments of DNA bound to specific transcription factors. But uncovering the second factor affecting binding (the DNA's 3D shape) has been no small task.
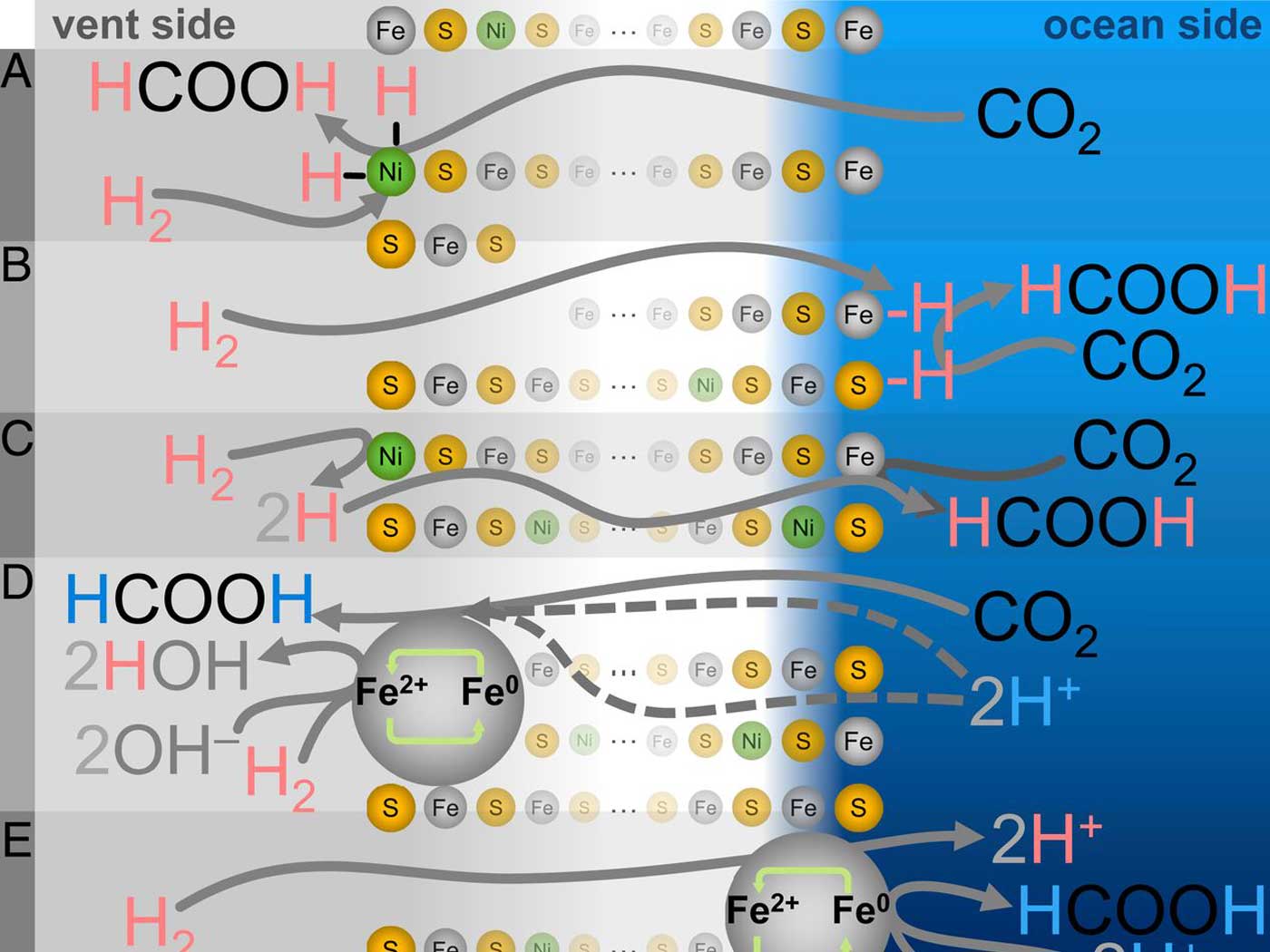
DNA is a double-stranded string of four different molecules called nucleotide bases (represented by the letters A, T, G, and C). The interdependence between nucleotide positions stems from the physical interactions between base pairs. These interactions give rise to the 3D structure of DNA which in turn helps transcription factors recognize specific sites for binding.2 Researchers in the present study observe that, "To date, a systematic and comprehensive survey of the value of shape-based models of DNA recognition has been lacking."1
The scientists used extensive data from large studies that assessed the protein binding biochemistry of transcription factors to DNA, as well as information gained from new biochemical investigations. They also had to employ a complex system of algorithm development using a technique called statistical machine learning to assess all the data. All three sources of information enabled them to develop a new binding model—one that considered DNA shape in addition to DNA sequence. When they tested their model against the standard model of using DNA sequence by itself, the new system greatly outperformed the standard model.
Past news reports have shown that multiple genome codes were contained in the same linear stretch of DNA sequences and accomplished different functions.3,4 Now we can add yet another type of code that is not based solely on DNA sequence, but rather the actual 3D shape of the DNA. A linear code plus a 3D code work together to tell the gene expression machinery precisely where to bind, turning genes off and on.
Yet another noteworthy feature of this new report is the fact that the failed theory of evolution was not given any credit whatsoever for accomplishing any of this amazing complexity. Obviously, to do so would have been utterly absurd—a type of oxymoron. The clear deduction from this surprising discovery—which required a combination of complex biochemistry and interpretive computer algorithms—is that an omnipotent Creator engineered it all in the beginning.
References
- Zhou, T., et al. 2015. Quantitative modeling of transcription factor binding specificities using DNA shape. Proceedings of the National Academy of Sciences. 112 (15): 4654–4659.
- Rohs, R., et al. 2009. The role of DNA shape in protein-DNA recognition. Nature. 461 (7268): 1248–1253.
- Tomkins, J. 2014. Duons: Parallel Gene Code Defies Evolution. Creation Science Update. Posted on icr.org January 6, 2014, accessed April 14, 2015.
- Tomkins, J. 2014. Dual-Gene Codes Defy Evolution...Again. Creation Science Update. Posted on icr.org September 12, 2014, accessed April 14, 2015.
*Dr. Tomkins is Research Associate at the Institute for Creation Research and received his Ph.D. in Genetics from Clemson University.
Article posted on April 27, 2015.